acespicoli
Well-known member
The orig SeedBanks on seedfinder (short list)
| Plankton |
|---|
| Part of a series on |

|
| show Trophic mode |
| show By size |
| hide By taxonomy |
| show By habitat |
| show Other types |
| show Blooms |
| show Related topics |







Welcomeawesome thread thanks

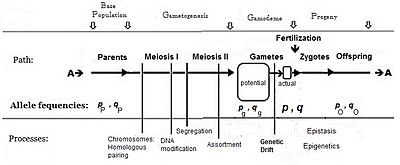
However, de Meijer et al. found that the THCA/CBDA ratio in medical marijuana F1 plants followed a Mendelian expectation of 1:2:1