acespicoli
Well-known member

When a gene is lost on the Y chromosome due to degeneration, it is essential for the organism's reproductive success that the corresponding function is still maintained. Therefore, through evolutionary processes, certain genes that were originally on the Y chromosome have been relocated to autosomes or the X chromosome.![The phylogeny is according to the reference of [59]. Idiograms created based on data obtained in [21], [23], [24], [26], [55] and in this study. 5S rDNA: green signals; 45S rDNA: red signals; species-specific subtelomeric repeats (HSR-1for H. lupulus, HJSR for H. japonicus and CS-1 for C. sativa): green signal. The position of pseudoautosomal region on sex chromosomes is indicated by brackets. Time of divergence estimated in [60], [61], [62]. The phylogeny is according to the reference of [59]. Idiograms created based on data obtained in [21], [23], [24], [26], [55] and in this study. 5S rDNA: green signals; 45S rDNA: red signals; species-specific subtelomeric repeats (HSR-1for H. lupulus, HJSR for H. japonicus and CS-1 for C. sativa): green signal. The position of pseudoautosomal region on sex chromosomes is indicated by brackets. Time of divergence estimated in [60], [61], [62].](https://www.researchgate.net/profile/Oleg-Aleksandrov/publication/259919693/figure/fig4/AS:202644292411425@1425325572964/The-phylogeny-is-according-to-the-reference-of-59-Idiograms-created-based-on-data.png)
The phylogeny is according to the reference of [59]. Idiograms created based on data obtained in [21], [23], [24], [26], [55] and in this study. 5S rDNA: green signals; 45S rDNA: red signals; species-specific subtelomeric repeats (HSR-1for H. lupulus, HJSR for H. japonicus and CS-1 for C. sativa): green signal. The position of pseudoautosomal region on sex chromosomes is indicated by brackets. Time of divergence estimated in [60], [61], [62].

An efficient RNA-seq-based segregation analysis identifies the sex chromosomes of Cannabis sativa
Cannabis sativa–derived tetrahydrocannabinol (THC) production is increasing very fast worldwide. C. sativa is a dioecious plant with XY Chromosomes, and only females (XX) are useful for THC production. Identifying the sex chromosome sequence would ...www.ncbi.nlm.nih.gov
Mapping of these sex-linked genes to a C. sativa genome assembly identified the largest chromosome pair being the sex chromosomes. We found that the X-specific region (not recombining between X and Y) is large compared to other plant systems. Further analysis of the sex-linked genes revealed that C. sativa has a strongly degenerated Y Chromosome and may represent the oldest plant sex chromosome system documented so far. Our study revealed that old plant sex chromosomes can have large, highly divergent nonrecombining regions, yet still be roughly homomorphic.
ONE DETAIL ID LIKE TO DISCUSS IF YOUR FOLLOWING IS
strongly degenerated Y Chromosome, AND WHY ?
They made a term "degeneration" and to me I see it as refinement or evolving towards perfectionWhen a gene is lost on the Y chromosome due to degeneration, it is essential for the organism's reproductive success that the corresponding function is still maintained. Therefore, through evolutionary processes, certain genes that were originally on the Y chromosome have been relocated to autosomes or the X chromosome.
I assume it can be called rapid degeneration cuz cannabis can lose gene snippets easily...very malleable plant within a few generations, new plant. The heteromorphy they talk about correlates to rapid degeneration in Y chrom cuz the plant wants to live super bad...it doesn't want to evolve into a new shape,form, structure, variant, so it replaces code quick and easy...they will not heteromorphy easily because they can repair chromosome nonrecombining regions. Just cuz it's quickest in the 3 species group ain't no big deal...except if you look at it from the point of view that this plant ain't supposed to dissappear or change dramatically ( wink)



There are two ways to perform selection; by looking at the plant and by looking at the DNA. In marijuana cultivation, plants are usually selected by looking at the plants.Only tiny differences in DNA are required to create major differences in appearance, growth or smoking effects.

But obviously it has to be decided which part of the hereditary information has to be used at a certain time and in a certain cell.All cells in the plant have the same genes. Each cell therefore has the same hereditary information and has the instructions to make each substance the plant can produce.
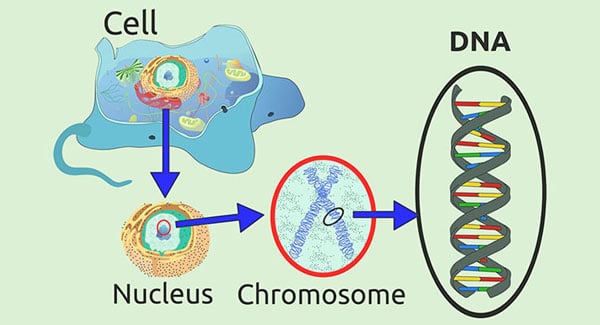
The order of bases in the DNA determines the order of chains of amino acids proteins are made of.The RNA leaves the nucleus of the cell and moves on to the ribosomes, where the code is read and the proteins are produced.

In Europe it’s not allowed to grow genetically modified plants, but this is legal for certain types of plant in other countries, such as the United States.For instance by adding a gene of a different plant species into the DNA of a certain plant. It’s also possible to put animal genes in plants or humans.





Cross | Flowering type | XX/XY | Flowering type | XX/XY | Germination rate (%) | Pure Male | Pure Female | MM |
|---|---|---|---|---|---|---|---|---|
| Pollen Donor | Ovule producer | Sexual phenotype of progeny (n=100) | |||||
1 | MM* | XY | Dioecious female* | XX | 84 | 0 | 35 | 49 |
2 | MM* | XY | MM* | XY | 42 | 0 | 24 | 18 |
3 | Dioecious female* | XX | Dioecious female* | XX | 78 | 0 | 78 | 0 |
4 | MM* | XY | Dioecious female+ | XX | 68 | 30 | 38 | 0 |
5 | Dioecious male+ | XY | Dioecious female* | XX | 60 | 21 | 36 | 3 |

| GENE | HGVS.C | HGVS.P | ANNOTATION | ANNOTATION IMPACT | CONTIG | CONTIG POS | REF/ALT | VAR FREQ |
|---|---|---|---|---|---|---|---|---|
| THCAS | c.749C>A | p.Ala250Asp | missense variant | moderate | contig741 | 4417079 IGV: Start, Jump | G/T | NGS: 0.127 C90: 0.632 |






In mapping studies the offspring that have alleles arranged as in the original parents are non-recombinants. |