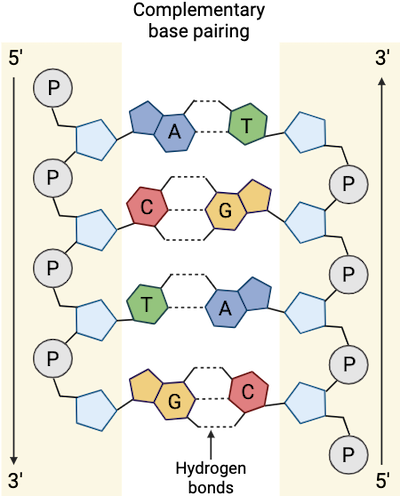
Male cannabis plants do serve an evolutionary purpose
Cannabis and most other species in the Cannabaceae family are dioecious, with males and females existing as separate plants (very few flowering plants possess this characteristic). The vast majority (over 80%) of flowering plants are hermaphroditic, meaning that each individual flower is comprised of both male and female sexual organs.Monoecy (where separate male and female flowers exist on the same plant) and dioecy are both rare, each comprising around 7% of flowering plant species. The remnant is made up of variations or mixtures of the three main types, (namely gynomonoecy, andromonoecy and trimonoecy) —where plants express both hermaphroditic flowers and female or male, respectively.

It is thought that dioecy confers a selective advantage in certain plant populations as it maximises the chances of genetic recombination. In hermaphrodite or monoecious plants, both male and female sexual organs are produced by the same plant. If it self-pollinates it will produce offspring whose DNA is identical to the parent. This lack of variation can very quickly lead to inbreeding and weakened genetic health in a population.
However, many hermaphroditic species have built-in genetic mechanisms that preclude self-pollination, a condition known as self-incompatibility.
It appears that monoecy and dioecy evolve in plants if the genetic mechanism for self-incompatibility has been lost (although they are each only present in around 7% of species, the capability has evolved independently in around 38% of all genera). There are several examples of dioecious plants expressing monoecious phenotypes in response to environmental pressures. But where those populations have again spread to more favourable locations, they gradually tend more towards dioecy.
This is an effective mechanism to ensure cross-pollination and genetic diversity in the absence of self-incompatibility.
This is borne out by cannabis, which has several monoecious strains and a great tendency to produce monoecious plants in dioecious populations, particularly in times of stress, and is entirely capable of self-pollination. In cannabis, numbers of males can drastically diminish for short periods in adverse conditions, but a strong and healthy male population is the default method to ensure long-term health and viability of the species.