-
ICMag with help from Landrace Warden and The Vault is running a NEW contest in November! You can check it here. Prizes are seeds & forum premium access. Come join in!
You are using an out of date browser. It may not display this or other websites correctly.
You should upgrade or use an alternative browser.
You should upgrade or use an alternative browser.
Phylos Galaxy - Landrace discussion
- Thread starter Thule
- Start date
@ngakpa
more than likely
with the DNA evidence on phylos it's pretty hard to dispute.
I'd be interested to see how qualified statisticians or botanists regard the data and system.
I strongly doubt either would regard Phylos in its current form as a sound basis for drawing firm conclusions about anything, least of all conclusions that are 'hard to dispute'
For example:
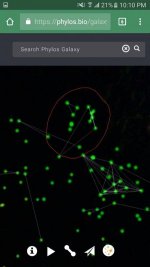
Could you explain why Senegal (West Africa) and Yarkhun (Chitral Hindu Kush) appear to be clustering together closer than Yarkhun and Laspur (the two valleys of Upper Chitral)?
I was with Mriko in Pakistan during one of his collecting trips to Chitral. You can read his public correspondence with Skunkman here on icmag in which he discusses giving the seeds to him.
As I say, Mriko's Yarkhun accession was acquired in Mastuj, which is the area at the junction of Yarkhun and Laspur Valleys in Upper Chitral.
His Laspur accession most likely was made in Sor Laspur, but if not then can only have been closer to Mastuj.
The maximum distance between the two landrace accessions from Chitral (Laspur Gold and Yarkhun) is 30km.
More to the point, Chitralis I spoke to regard these as being accessions of the same landrace. The plants look the same, including the seeds. The resin smells, looks, and smokes the same. Between Sor Laspur and Mastuj is a continuous run of villages in which people use and share the same seeds!
Based on my admittedly limited understanding of statistics, the numbers you are working with for any one landrace (Colombian Gold) or hybrid (Haze) simply aren't adequate to make any confident assertions.
And there are a host of questions regarding genetics in cultivars and cultigens that bear on the interpretation of these diagrams, particularly as regards the effect of breeding on genetic profiles, that require specialist botanical and genetic knowledge in order to interpret the significance of the system.
At the minute, the Phylos system looks to be like another trap for the kind of typological thinking that has already caused so much confusion in cannabis.
(Even the whole NLD/BLD thing is dodgy btw and based on misunderstandings about domestication)
brickweeder
Well-known member
...or this concept is fail...maybe better they did family "tree-root" marked branching like on Seed.finder
Generally Agree. For me, S#ed###der is way more useful than phylos, and it doesn't crash/lock my browser like phylos does. Plus seed-fine-der lets me easily weed through all the BS marketing strain names to see what was bred with what and to be able to ID the underlying parent races and source geography.
I'd be interested to see how qualified statisticians or botanists regard the data and system.
I strongly doubt either would regard Phylos in its current form as a sound basis for drawing firm conclusions about anything, least of all conclusions that are 'hard to dispute'
For example:
Could you explain why Senegal (West Africa) and Yarkhun (Chitral Hindu Kush) appear to be clustering together closer than Yarkhun and Laspur (the two valleys of Upper Chitral)?
I was with Mriko in Pakistan during one of his collecting trips to Chitral. You can read his public correspondence with Skunkman here on icmag in which he discusses giving the seeds to him.
As I say, Mriko's Yarkhun accession was acquired in Mastuj, which is the area at the junction of Yarkhun and Laspur Valleys in Upper Chitral.
His Laspur accession most likely was made in Sor Laspur, but if not then can only have been closer to Mastuj.
The maximum distance between the two landrace accessions from Chitral (Laspur Gold and Yarkhun) is 30km.
More to the point, Chitralis I spoke to regard these as being accessions of the same landrace. The plants look the same, including the seeds. The resin smells, looks, and smokes the same. Between Sor Laspur and Mastuj is a continuous run of villages in which people use and share the same seeds!
Based on my admittedly limited understanding of statistics, the numbers you are working with for any one landrace (Colombian Gold) or hybrid (Haze) simply aren't adequate to make any confident assertions.
And there are a host of questions regarding genetics in cultivars and cultigens that bear on the interpretation of these diagrams, particularly as regards the effect of breeding on genetic profiles, that require specialist botanical and genetic knowledge in order to interpret the significance of the system.
At the minute, the Phylos system looks to be like another trap for the kind of typological thinking that has already caused so much confusion in cannabis.
(Even the whole NLD/BLD thing is dodgy btw and based on misunderstandings about domestication)
Some lab tech was getting high on some old nld he was supposed to be running tests on and mislabeled an entire weeks worth of work. Not much different than labeling your favorite strain wrong and not noticing in time.
Just kidding.........
Trash
frostqueen
Active member
Image anti-clockwise from top:
European hemp
late 70's American hybrids/central american
Mr Nice Haze/NYC Haze
Colombian Gold
Colombian Red
Thai Stick/Asia Sativa
African Sativas
Cool... circled colored dots. I'm confused as to why you are posting these. What exactly are you trying to show us with all of these posts?
Nexus7
Well-known member
@ngakpa
Well I'm pretty confident the phylos system works well and there is enough data there already to start drawing conclusions ....
I'd even be happy to wager you the cost of a pack of your Kerala seeds that if you were to submit it that it would fall next to the other Kerala samples submitted in the cluster I marked (southern) India just adjacent to the Thai/Asia cluster I have market.
While I can't explain away everything just yet some strange things are likely to have occured in regard to Canbabis transport in the past. Also I think Cannibis is one of those things where environment plays a major part on the expression of the genes. Much more than people currently believe.
Well I'm pretty confident the phylos system works well and there is enough data there already to start drawing conclusions ....
I'd even be happy to wager you the cost of a pack of your Kerala seeds that if you were to submit it that it would fall next to the other Kerala samples submitted in the cluster I marked (southern) India just adjacent to the Thai/Asia cluster I have market.
While I can't explain away everything just yet some strange things are likely to have occured in regard to Canbabis transport in the past. Also I think Cannibis is one of those things where environment plays a major part on the expression of the genes. Much more than people currently believe.
Nexus7
Well-known member
Cool... circled colored dots. I'm confused as to why you are posting these. What exactly are you trying to show us with all of these posts?
So what I'm trying to show people is that regions or clusters of samples are forming that correspond to geographical regions.
So if you have a legit Thai-stick and you were to submit it it would fall in the region I circled Asia/Thai.
Also say you have a top Sativa you are unsure of it's origins you submit it to phylos and in fell in the cluster I marked Africa. You have an African Sativa my friend.
So simple it's hard to comprehend I know
Nexus7
Well-known member
@ Dog Star and brickweeder
While I did enjoying looking a s**dfinder and have found it useful in the past a lot of the sample lineages are based on best guess. DNA is the future because it can prove 100% who parents what.
Unfortunately a lot of breeders are mistaken as to what they have got and some will just plain lie about provedence and some others just like to keep it a secret.
Nothing against euseedfind I found it very useful previously but it's just a starting place for me and DNA evidence is needed to settled disputed lineages imo.
While I did enjoying looking a s**dfinder and have found it useful in the past a lot of the sample lineages are based on best guess. DNA is the future because it can prove 100% who parents what.
Unfortunately a lot of breeders are mistaken as to what they have got and some will just plain lie about provedence and some others just like to keep it a secret.
Nothing against euseedfind I found it very useful previously but it's just a starting place for me and DNA evidence is needed to settled disputed lineages imo.
@ngakpa
Well I'm pretty confident the phylos system works well and there is enough data there already to start drawing conclusions ....
I'd even be happy to wager you the cost of a pack of your Kerala seeds that if you were to submit it that it would fall next to the other Kerala samples submitted in the cluster I marked (southern) India just adjacent to the Thai/Asia cluster I have market.
While I can't explain away everything just yet some strange things are likely to have occured in regard to Canbabis transport in the past. Also I think Cannibis is one of those things where environment plays a major part on the expression of the genes. Much more than people currently believe.
in fairness, if you're who I suspect you may be, you already know that I have already offered OCP (who are affiliated with Phylos) a selection of seeds of each of the RSC strains for free
and if not, then you do now
we've got two options here:
1. accept that these anomalies indicate that there are fundamental questions about the technique and hermeneutics
2. assert that 'some strange things are likely to have occured in regard to Canbabis transport in the past'
let's just say that ideas such as that Yarkhun seed found its way to Casamance or Panama Red was created from hemp and a North Indian feral landrace are not things I'd be wanting to shout about if I was involved with Phylos
Also I think Cannibis is one of those things where environment plays a major part on the expression of the genes. Much more than people currently believe.
right, but that's just one example of several fundamental questions that require the specialist involvement of geneticists and botanists
DNA evidence is needed to settled disputed lineages imo.
I don't think anyone is denying this
but DNA-based evidence is one thing
the ability to interpret DNA-based evidence is another
in the absence of specialist knowledge on genetics and botany for cannabis, you've got major hermeneutic question marks hanging over this whole thing
plus there's the issue of statistically significant data
at the minute, this is just taking you down the same road of typological confusion that cannabis has been on since Schultes
So what I'm trying to show people is that regions or clusters of samples are forming that correspond to geographical regions.
So if you have a legit Thai-stick and you were to submit it it would fall in the region I circled Asia/Thai.
Also say you have a top Sativa you are unsure of it's origins you submit it to phylos and in fell in the cluster I marked Africa. You have an African Sativa my friend.
So simple it's hard to comprehend I know
pride comes before a fall
I suggest you head off and familiarise yourself with statistics and typology, particularly as regards how typological thinking has created mayhem in cannabis taxonomy for most of the past five decades
you're going to need a load more samples before you can be so confident
Nexus7
Well-known member
pride comes before a fall
I suggest you head off and familiarise yourself with statistics and typology, particularly as regards how typological thinking has created mayhem in cannabis taxonomy for most of the past five decades
you're going to need a load more samples before you can be so confident
Well it looks like we will have to agree to disagree then. Although I concede the current data on phylos is still in it's infancy I still think we can come up with some theories and draw some conclusions already.
Naturally as more samples are submitted current working theories in regard to Cannabis evolution and speciation will be confirmed, refuted or needed to be modified. There's no harm in early speculation imo given the current state of affairs.
As I mentioned earlier I'm more than happy to put my pride, ego and money etc on the line in regards to the Kerala you collected and it's likely place in the phylos galaxy.
Are you willing to do the same and test my understanding of the phylos database?
Nexus7
Well-known member
in fairness, if you're who I suspect you may be, you already know that I have already offered OCP (who are affiliated with Phylos) a selection of seeds of each of the RSC strains for free
and if not, then you do now
we've got two options here:
1. accept that these anomalies indicate that there are fundamental questions about the technique and hermeneutics
2. assert that 'some strange things are likely to have occured in regard to Canbabis transport in the past'
let's just say that ideas such as that Yarkhun seed found its way to Casamance or Panama Red was created from hemp and a North Indian feral landrace are not things I'd be wanting to shout about if I was involved with Phylos
right, but that's just one example of several fundamental questions that require the specialist involvement of geneticists and botanists
Sorry I missed this post. No I don't work or am affiliated in any way with Phylos, OCP or any such companies.
I don't care if you do or don't submit your collections to phylos or kannapedia or wherever else.
I just offered to test your kerala sample to prove a point. It only has one possible place in the phylos galaxy and that is with all the other kerala's otherwise it wouldn't be Kerala would it? Assuming that when we talk of Kerala we are talking about the select NLD/tropical Sativa cultivar and not the whole
region of Kerala where anyone is free to grow whatever they wish.
Yes in hindsight the Panama Red theory I mentioned earlier is pretty laughable but in all fairness I was pretty high at the time ...
I guess you're more concerned with population genetics of a region at a particular point in time whereas I'm more interested in the selections of those genetics that produce a uniform variety that shares a common set of phenotypic traits and DNA. Sure the genetics will drift over time but they will always share a similar signature as long as no new genetics are introduced.
Also it's never really going to be feasible to genotype 1000 or more individuals from a particular region to find out the level of variation at the population level.
Remember what the endgame of phylos is after all?
Please take my money and test my OG Kusk/Cookies/Gg4 etc and tell me if it's real deal or not ...
Sorry I missed this post. No I don't work or am affiliated in any way with Phylos, OCP or any such companies.
I don't care if you do or don't submit your collections to phylos or kannapedia or wherever else.
I just offered to test your kerala sample to prove a point. It only has one possible place in the phylos galaxy and that is with all the other kerala's otherwise it wouldn't be Kerala would it? Assuming that when we talk of Kerala we are talking about the select NLD/tropical Sativa cultivar and not the whole
region of Kerala where anyone is free to grow whatever they wish.
Yes in hindsight the Panama Red theory I mentioned earlier is pretty laughable but in all fairness I was pretty high at the time ...
I guess you're more concerned with population genetics of a region at a particular point in time whereas I'm more interested in the selections of those genetics that produce a uniform variety that shares a common set of phenotypic traits and DNA. Sure the genetics will drift over time but they will always share a similar signature as long as no new genetics are introduced.
Also it's never really going to be feasible to genotype 1000 or more individuals from a particular region to find out the level of variation at the population level.
Remember what the endgame of phylos is after all?
Please take my money and test my OG Kusk/Cookies/Gg4 etc and tell me if it's real deal or not ...
In fairness, I've worked in academic copy editing for the sciences, which means I've read way more papers on statistics than any person should ever have to read
I can assure you, even with my limited understanding of statistics that you are getting way ahead of yourself
I'll happily send you a pack of Kerala if you're (1) not in the US and (2) actually going to pay for the DNA profile (which Phylos themselves won't, they merely collate the data)
when we talk of Kerala we are talking about the select NLD/tropical Sativa cultivar and not the whole
region
it's worth noting that the best term to use for a domesticated landrace is cultigen
for the sake of clarity, 'cultivar' is probably best reserved for formally recognised, registered cultivars
this use of this term cultivar by some cannabis experts to apply to domesticated landraces is problematic
the same applies wih using cultivar to apply to most modern Western drug 'strains', which are largely a mess of random hybrids, nothing approaching the level of unformity and stability required for a recognised cultivar
formal cultivars and landraces are very different things
the fundamental point about landraces is their vast range of variation and genetic diversity - it's really the crucial point here
there's a recognisable Idukki landrace, but there's a great deal of inherent and potential genetic and phenotypic variation within that - vastly more than there is with a formal cultivar or a modern slop hybrid
I was watching a 2015 panel discussion on this just now, where our anointed cannabis experts were talking as if 'Thai' is one uniform landrace, and as if Afghan landraces are descended from one primordial 'afghanica' ancestor... silly talk, really...
I still think we can come up with some theories and draw some conclusions already.
you can have as many theories and draw as many conclusions as you want
the question is whether they have any value or merit